Research
A full list of my publications are available here: Google Scholar
Efficient Modeling of Polygenic Risk Scores and Environment Interplay
We developed a series of novel methods for modeling of polygenic scores (PGS) in different epidemiological study designs. These include joint modeling of polygenic gene-environment correlations and interactions in case-control studies, and unified modeling of direct, indirect, and gxe interactions in family-based studies. We have shown improved efficiency in case-control studies that are comparable to cohort studies. Family-based methods can further eliminate potential population stratification bias in PGS estimations.
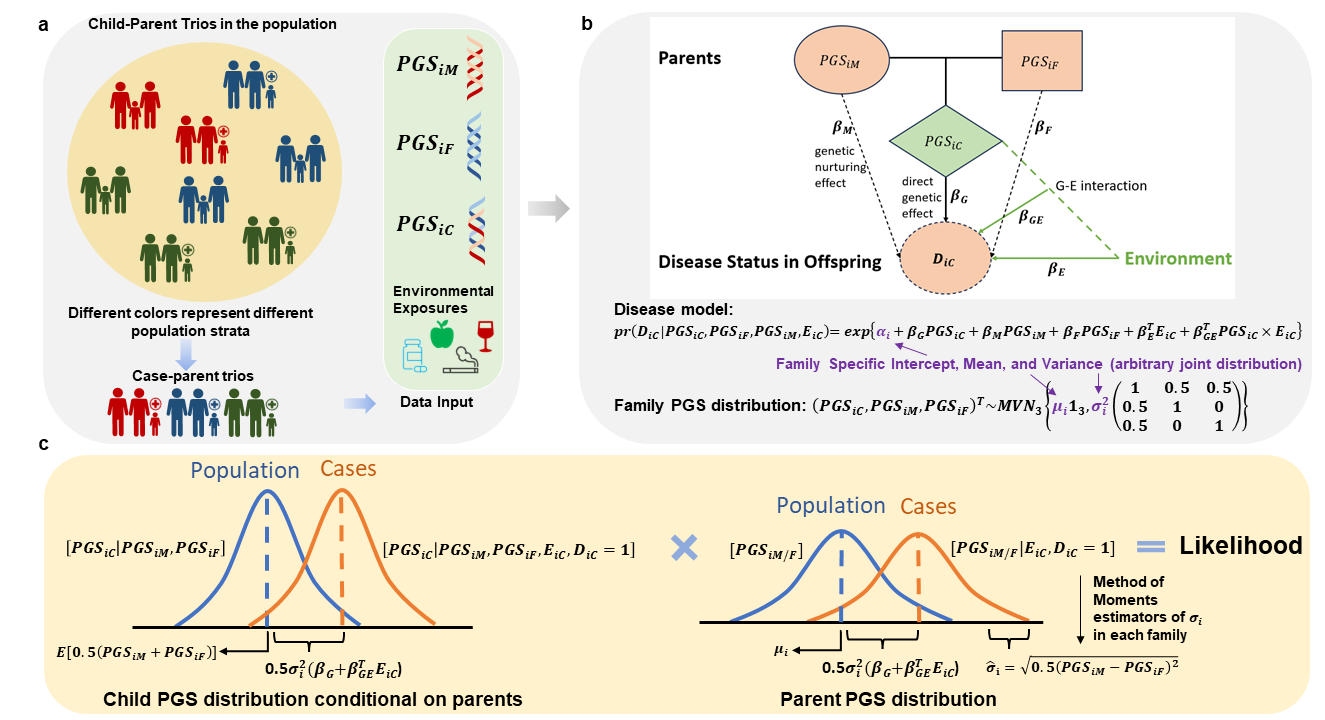
- Wang, Z., Shi, W., Carroll, R., Chatterjee, N. (2024). Joint Modeling of Gene-Environment Correlations and Interactions Using Polygenic Risk Scores in Case-Control Studies. American Journal of Epidemiology. DOI: 10.1093/aje/kwae081 [Paper] [Software]
- Wang, Z., Grosvenor, L., Ray, D., Ruczinski, I., Beaty, T., Volk, H., Ladd-Acosta, C., Chatterjee, N. (2024+). Estimation of Direct and Indirect Polygenic Effects and Gene-Environment Interactions using Polygenic Scores in Case-Parent Trio Studies. [Preprint] [Software]
Multi-Omics Data Integration
We developed novel methods for data integrations to examine the associations between genes and disease phenotype through multiple layers of omic information. Our methods are based on multivariate Gaussian mixture models to characterize the precise correlations between different data types using summary statistics, leading to superior statistical power in association studies. We have also extended this work to spatially resolved high-dimensional genomics data, including DNA methylation and RNA sequencing data using specific datasets in bladder tumor samples, to increase the power of finding molecular signals contributing to cancer initiation, promotion, and progression.
Our work has several key advantages over conventional designs, including the novel feature of controlling the false discovery rate in significant genes across multiple omics data types, the ability to incorporate independent or overlapping sets of cohorts with respect to omics data, the generality of applications to meta-analysis and gene enrichment pathways, and fast computational efficiency by adopting the expectation-maximization (EM) algorithm.
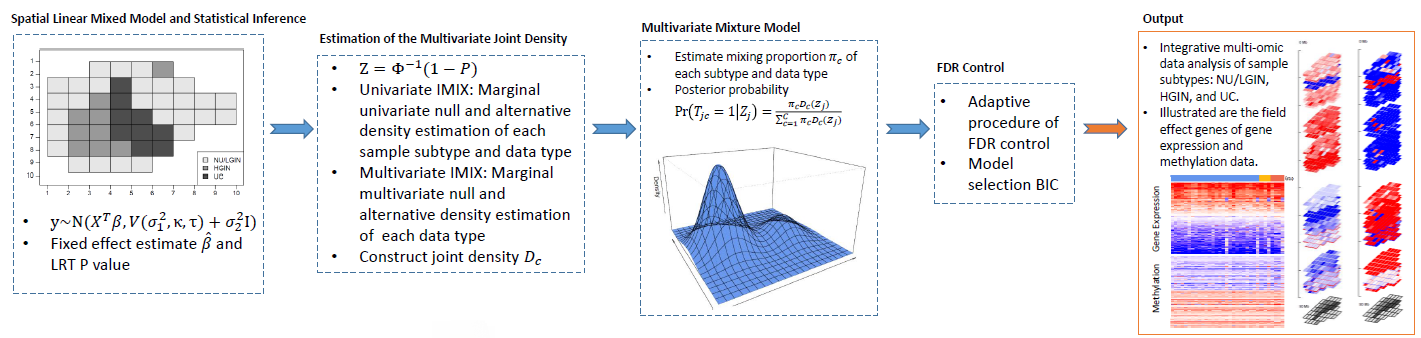
- Wang, Z., Wei, P. (2020). IMIX: A Multivariate Mixture Model Approach to Association Analysis Through Multi-Omics Data Integration. Bioinformatics. DOI:10.1093/bioinformatics/btaa1001 [Paper] [Software] (Platform presentation, ASHG 2020)
- Wang, Z., Czerniak, B., Wei, P. (2023). Spatial IMIX: A Mixture Model Approach to Spatially Correlated Multi-Omics Data Integration. Preprint. DOI:10.1101/2023.07.15.549148 [Paper] [Software]
Epigenome-Wide Association Studies in Cancer
We also investigated the role of DNA methylation in whole blood affecting susceptibility to pancreatic cancer. We performed a two-phase epigenome-wide association study by examining two independent case-control studies measured within MDA and leveraged the Framingham Heart Study data to increase the sample size in controls. We discovered important differentially methylated CpG sites and regions associated with pancreatic cancer, and validated the results using a third independent dataset from the Women’s Health Initiative. We further confirmed the causal relationship of the discovered CpG sites using Mendelian randomization analysis, and investigated the functional consequences in gene expression. We performed another large-scale EWAS in studying the role of differentially methylated CpG sites between HPV-positive and HPV-negative oropharyngeal cancer patients and discovered a long non-coding RNA nc886 that leads to further important clinical implications.
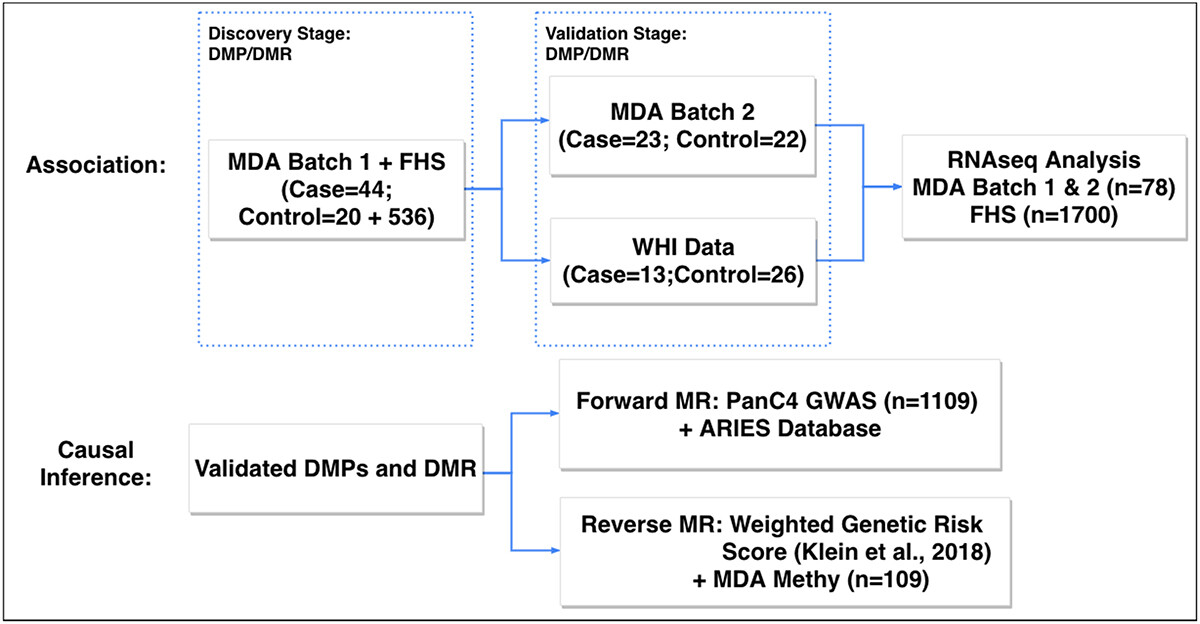
- Wang, Z., Lu, Y., Fornage, M., Jiao, L., Li, D., Wei, P. (2022). Identification of novel susceptibility methylation loci for pancreatic cancer in a two-phase epigenome-wide association study. Epigenetics. DOI: 10.1080/15592294.2022.2026591 [Paper] (Platform presentation, IGES 2019)
- Xu, Y.(co-first author), Wang, Z.(co-first author), Wei, P., Gairola, R., Kelsey, K., Sikora, A., Li, G., Gu, J. (2022). Hypermethylation of nc886 in HPV-positive oropharyngeal cancer and its clinical implications: an epigenome DNA methylation profiling study. Molecular Therapy-Nucleic Acids. DOI:10.1016/j.omtn.2022.11.012 [Paper]
Method for Benefit-Risk Analysis in Clinical Trials
I collaborated with Dr. Jie Chen at Merck during my summer internship and developed a novel method to test for significant trend effects of multiple dosage levels of medication use by using composite endpoints in clinical trial studies. This method prioritizes the user’s intended endpoints including the adverse events and efficacy endpoints and assesses whether the examined dose levels in clinical trials show an increasing or decreasing trend effect. This method is computationally fast and robust to type I errors.
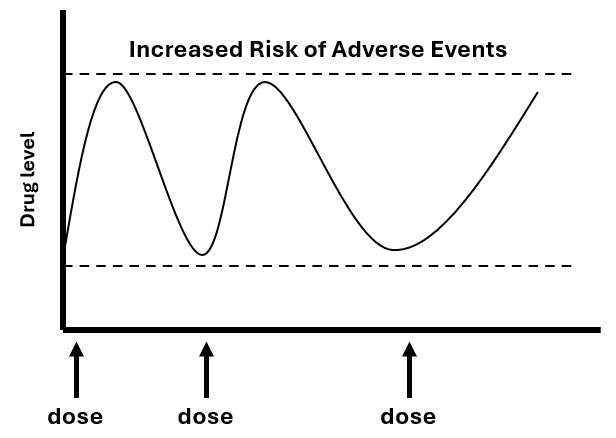
- Wang, Z., Chen J. Testing for Trend in Benefit-Risk Analysis with Prioritized Multiple Outcomes. (2020). Statistics in Biopharmaceutical Research. DOI: 10.1080/19466315.2019.1690037 [Paper] [Software]
Collaborations in Biomedical Sciences
I welcome collaborations with biologists, physicians, epidemiologists, and other biomedical researchers. I did extensive collaboration work with biomedical researchers at MD Anderson Cancer Center during my PhD study. Applying my knowledge in statistical genetics and genomics, I supported a variety of important research work in molecular biology studying bladder cancer, pancreatic cancer, and head and neck cancer.
- Bondaruk, J.(co-first author), Jaksik, R.(co-first author), Wang, Z.(co-first author), Cogdell, D.(co-first author), Lee, S., Chen, Y., Dinh, K., Majewski, T., Zhang, L., Cao, S., Yao, H., Weinstein, J., Navai, N., Dinney, C., Gao, J., Theodorescu, D., Logothetis, C., Guo, C., Wang, W., McConkey, D., Wei, P., Kimmel, M., Czerniak, B. (2022). The origin of bladder cancer from mucosal field effects. iScience. DOI: 10.1016/j.isci.2022.104551 [paper]
- Yang, G.(co-first author), Bondaruk, J.(co-first author), Cogdell, D.(co-first author), Wang, Z.(co-first author), Lee, S., Lee, J., Zhang, S., Choi, W., Wang, Y., Liang, Y., Wang, G., Wang, Y., Yao, H., Dadhania, V., Logothetis, C., Siefker-Radtke, A., Kamat, A., Dinney, C., Theodorescu, D., Kimmel, M., Wei, P., Guo, C., Weinstein, J., McConkey, D., Czerniak, B. (2020). Urothelial-to-Neural Plasticity Drives Progression to Small Cell Bladder Cancer. iScience. DOI: 10.1016/j.isci.2020.101201. [paper]
- Lee, S., Bondaruk, J., Wang, Y., Chen, H., Lee, J., Majewski, T., Mullen, R., Cogdell, D., Chen, J., Wang, Z., Yao, H., Kus, P., Jeong, J., Lee, I., Choi, W., Navai, N., Guo, C., Dinney, C., Baggerly, K., Mendelsohn, C., McConkey, D., Behringer, R., Kimmel, K., Wei, P., Czerniak, B. Loss of LPAR6 and CAB39L dysregulates the basal-to-luminal urothelial differentiation program, contributing to bladder carcinogenesis. (2024). Cell reports. DOI: 10.1016/j.celrep.2024.114146. [paper]
- Guo, C. C., Bondaruk, J., Yao, H., Wang, Z., Zhang, L., Lee, S., Lee, J. G., Cogdell, D., Zhang, M., Yang, G., Dadhania, V., Choi, W., Wei, P., Gao, J., Theodorescu, D., Logothetis, C., Dinney, C., Kimmel, M., Weinstein, J. N., McConkey, D. J., … Czerniak, B. (2020). Assessment of Luminal and Basal Phenotypes in Bladder Cancer. Scientific reports. DOI: 10.1038/s41598-020-66747-7. [paper]
- Guo, C., Majewski, T., Zhang, L., Yao, H., Bondaruk, J., Wang, Y., Zhang, S., Wang, Z., Lee, J., Lee, S., Cogdell, D., Zhang, M., Wei, P., Grossman, H., Kamat, A., Duplisea, J., Ferguson, J., Huang, H., Dadhania, V., Dinney, C., Weinstein, J., Baggerly, K., McConkey, D., Czerniak, B. (2019). Dysregulation of EMT Drives the Progression to Clinically Aggressive Sarcomatoid Bladder Cancer. Cell reports. DOI: 10.1016/j.celrep.2019.04.048. [paper]
- Zhang, Y., Sturgis, E.M., Wei, P., Liu, H., Wang, Z., Ma, Y., Liu, C., Gu, K.J., Wei, Q., and Li, G. A Genetic Variant within MDM4 3’UTR miRNA Binding Site is Associated with HPV16-positive Tumors and Survival of Oropharyngeal Cancer. (2019). Molecular carcinogenesis. DOI:10.1002/mc.23116. [paper]
- Yang, Z., Zhang, Y., Wang, X., Huang, J., Guo, W., Wei, P., Li, G., Wang, Z., Huang, Z., and Zhang, L. Putative Biomarkers of Malignant Transformation of Sinonasal Inverted Papilloma into Squamous Cell Carcinoma. (2019). The Journal of international medical research. DOI:10.1177/0300060519838385. [paper]
Currently at Johns Hopkins University, I am leading the effort in identifying important plasma proteins associated with common solid cancer incidence. I also performed Mendelian randomization analyses to identify potential causal relationships between proteins and incident hypertension. I have also supported projects constructing polygenic risk scores for type 2 diabetes and coronary artery disease, as well as unsupervised clustering using proteomics data to identify disease subtypes in stroke.
- Wang, Z., Burk, V., Chatterjee, N., Platz, E. (2024+). Discovering plasma proteins associated with common solid cancer incidence in ARIC.
- Mathews, L., Hu, X., Wang, Z., et al. (2024+). MMP7 as a Causal Factor for Incident Hypertension and its Complications Proteomic Analysis from the Atherosclerosis Risk in Communities (ARIC) Study and Mendelian Randomization Study.
- Dzaye, O., …, Wang, Z., Chatterjee, N., Matsushita, K., Blaha, M. (2024). Polygenic Risk Scores and Extreme Coronary Artery Calcium Phenotypes in Adults >75 Years Old: The Atherosclerosis Risk in Communities Study. Circulation: Cardiovascular Imaging. DOI: 10.1161/CIRCIMAGING.123.016377. [paper]
- Zhang, M., Ru, M., Zhang, J., Wang, Z., et al. (2024). Alcohol consumption does not modify the polygenic risk score-based genetic risk of breast cancer in postmenopausal women: Atherosclerosis Risk in Communities study. Cancer Prevention Research. DOI: 10.1158/1940-6207.CAPR-24-0208. [paper]